GVHD
CLASSIFICATION FOR GVHD AND TUBULAR ADENOMA
LABELING
In this task, coordinates of the marked region of interest (ROI) are extracted with the QuPath application. ROIs have been chosen according to the blue marked areas on the image and each blue marked has been labeled with different sizes and shapes to increase the data variation. Extracted coordinates are utilized on unmarked images to prevent blue marks exist on marked images.
For each image, around 40-50 ROIs are extracted. The center coordinate of each ROI is used as the base for cropping areas with different sizes of 128×128 and 256×256.
The ROIs are labelled in three class identifiers; 0. Tubular Adenoma, 1. GVHD, 2. None
The ROIs with ‘None’ labels are selected randomly based on the non-overlapping coordinates that exclude GVHD and Tubular Adenoma coordinates.
FEATURE EXTRACTION
Feature extraction is a method for generating numerical representations of an image, which can then be used as input for a model. The models known as foundational models are trained especially for this purpose.
-> [0.0010228882,0.821333,0.48481488,0.13029815]
The foundational model we used for feature extraction on GVHD images is ‘Prov-Gigapath’. This model generates a vector with a shape of 1×1536 floats.
For 728 different ROIs, the total features generated consist of 728 rows and 1536 columns within a CSV file.
IDENTIFYING AND CLASSIFICATION
The classification task utilized in our study aims to see whether generated features are distinguishable among the other classes. As it is mentioned there are 3 different classes that are labelled regarding the coordinates of ROIs. This label information was also added as a separate column in our CSV file as a ‘label’ column.
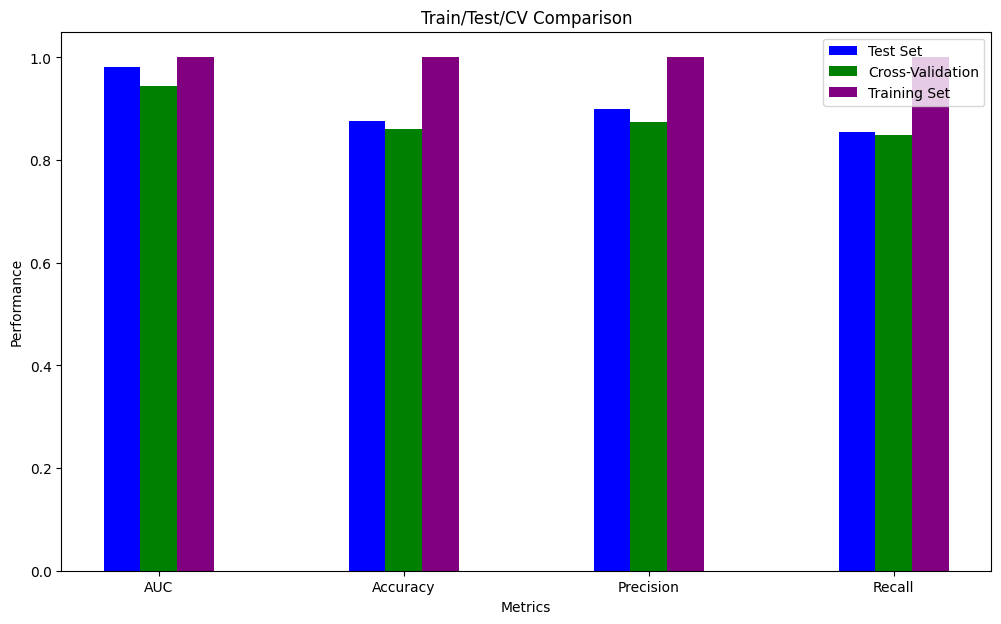
The classification experiment conducted on the random forest algorithm shows that the AUC (area under the curve), which represents the model’s ability to distinguish between classes, demonstrates the model’s performance. An AUC value closer to 1 indicates a better performing model, while a value closer to 0.5 suggests a model with no discriminative ability.
| dataset | model | test_auc | test_acc | test_precision | test_recall | test_kappa | cvt_auc | cvt_acc | cvt_precision | cvt_recall |
| gvhd_128.csv | randomforest | 0.982 | 0.877 | 0.9 | 0.854 | 0.796 | 0.944 | 0.86 | 0.874 | 0.849 |
You can also see the additional classification results in the image below: